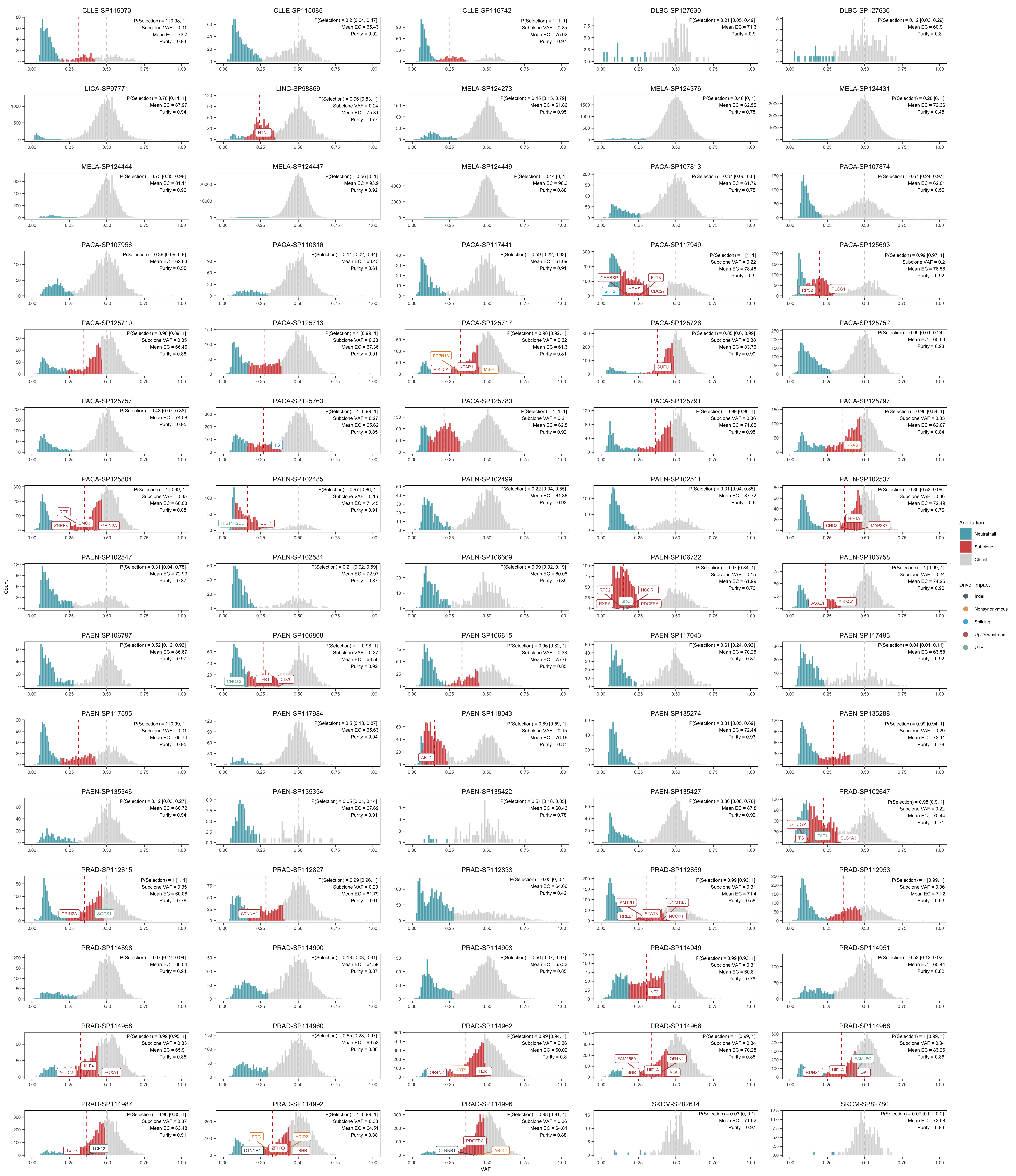
Supplementary Figure 19. VAF distributions with annotated TumE fits for 75 PCAWG samples with either zero or one detected subclone. Each tumour was classified as either neutrally evolving or subject to positive selection using TumE run with 250 Monte Carlo dropout samples. Samples subject to positive selection were further annotated as 1 or 2 subclone. This figure displays all TumE fits in samples classified as neutrally evolving or carrying 1 subclone. If a subclone was present, we annotated each plot with known driver mutations and the functional impact of the corresponding mutation. The statistics generated for the three samples identified with 2 subclones can be found in the Supplementary Tables associated with the paper.