
Supplementary Figure 17. Evaluation of the peak-finding/heuristic VAF adjustment method.We evaluated TumE estimates in 6000 tumours with and without correcting for tumour purity. Tumour purity ranged from 0.2 - 1 and sequencing depth ranged from 20 - 200. Our analyses suggest tumour purity correction taking into account clonal peak adjustment and diploid purity adjustment (i.e. VAF/purity) is necessary to ensure accurate prediction in low purity samples.
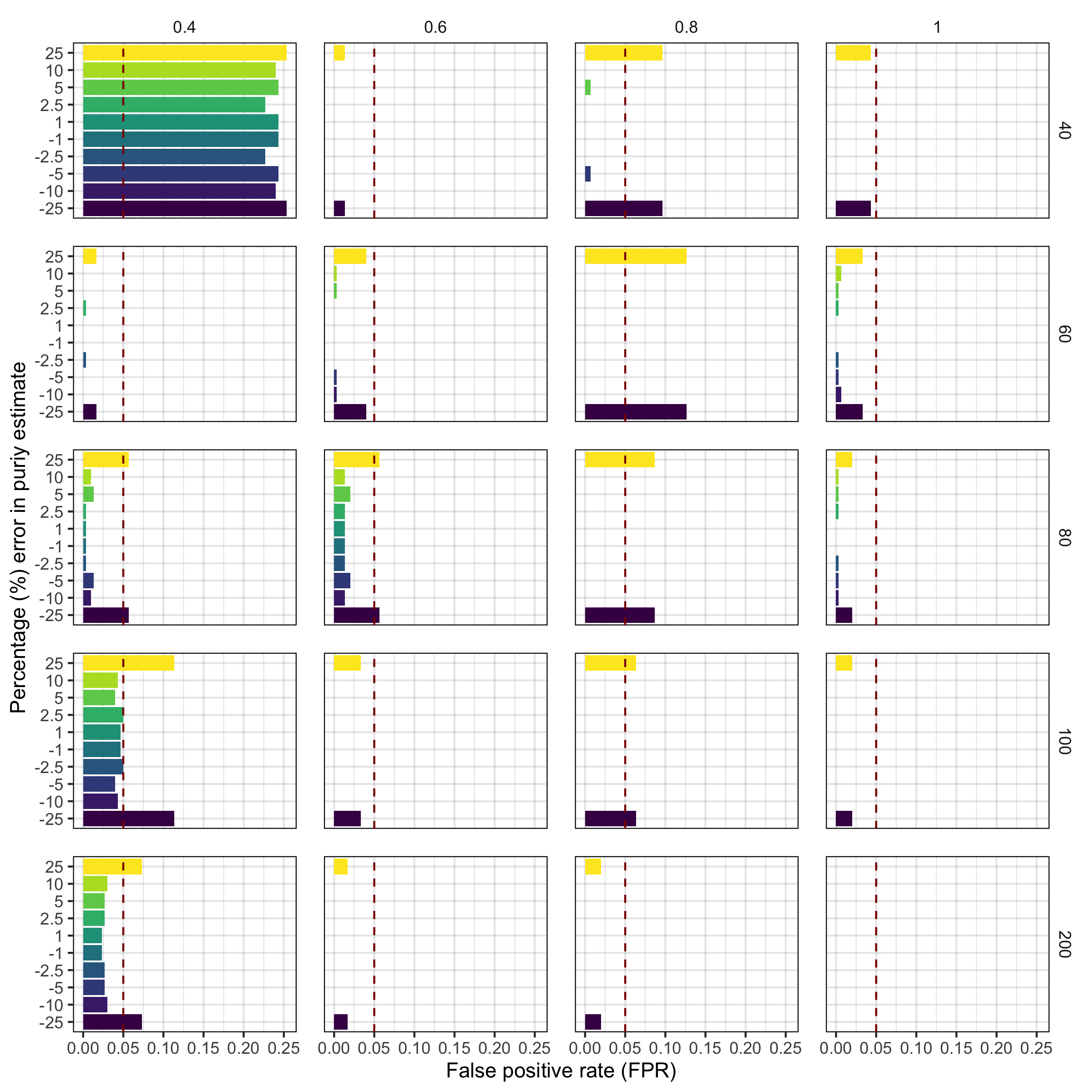
Supplementary Figure 18. False positive rate for positive selection (>= 1 subclone) at variable sequencing depths, tumour purities, and errors in purity estimates in 6000 synthetic tumours.We evaluated TumE estimates in 6000 synthetic tumours following the addition of different levels of incorrect purity estimates ranging from 1 to 25% above or below the true purity. VAFs from each synthetic tumour were corrected using the peak-finding/heuristic adjustment method described in Methods. We found TumE estimates using this adjustment were robust to errors in purity estimates up to 25%, maintaining a <5% false positive rate at mean effective sequencing depths of ~40x. The top labels on each panel column is the true tumour purity. The right labels on each panel row are the mean sequencing depth for each synthetic tumour.